Publication
A unified framework for integrative study of heterogeneous gene regulatory mechanisms
(Published, Nature Machine Intelligence)
Author: Qin Cao, Zhenghao Zhang, Alexander Xi Fu, Qiong Wu, Tin-Lap Lee, Eric Lo, Alfred S. L. Cheng, Chao Cheng, Danny Leung, and Kevin Y. Yip
What is GEEK?
GEEK (Gene Expression Embedding frameworK) is a flexible framework that can unify various heterogeneous biological data sources to perform integrative studies.
Current demonstrative GEEK pipeline supports integrating data types of:
- PPI (Protein-Protein Interaction): a static network in which each node is a gene and each edge is a physical interaction between two proteins encoded by the corresponding genes.
- Gene-bin association: a static network in which each node is either a gene or a genomic bin, and each edge connects a gene to all the genomic bins that overlap its locus.
- Chromatin accessibility: a cell type-specific numeric feature of the average chromatin accessibility of each genomic bin.
- Chromosome conformation: a cell type-specific network, in which each node is a genomic bin and each edge is a statistically significant interaction between two bins in the three-dimensional genome architecture, with the edge weight representing the significance of the interaction.
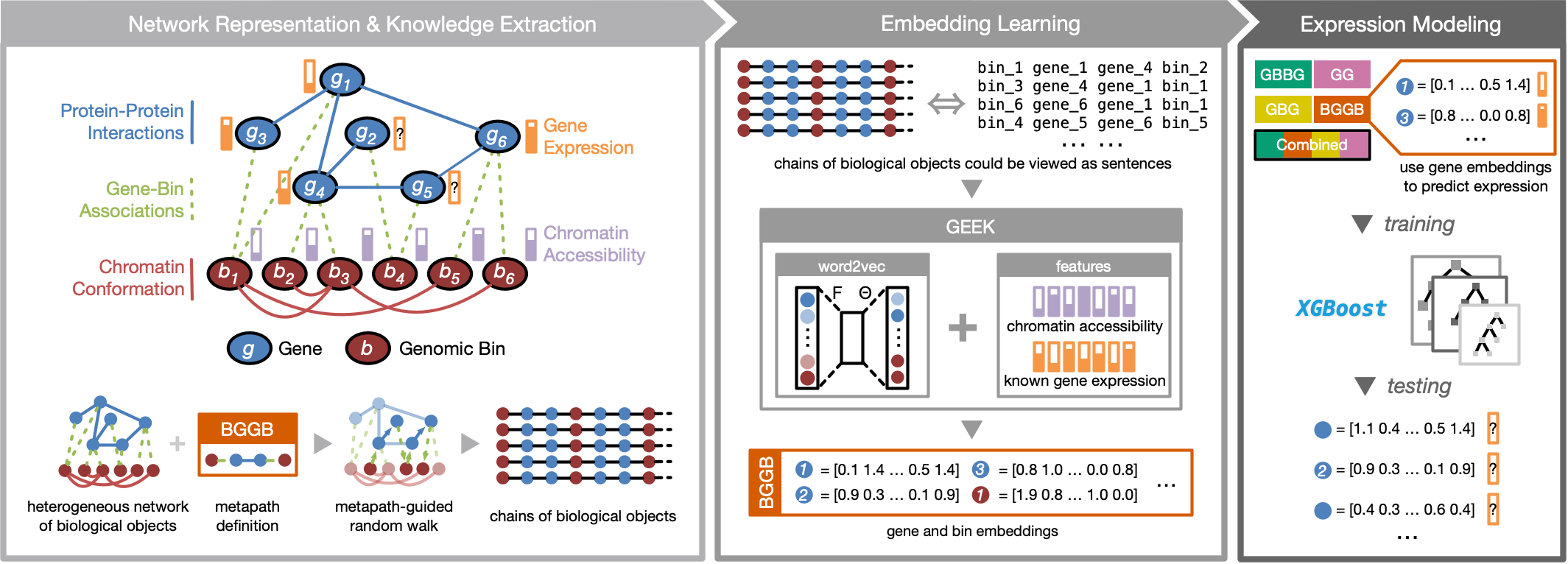
GEEK is powered by network embedding techniques originated from metapath2vec, which could learn topological information following predefined patterns (metapaths), along with our modifications to handle features on biological objects.
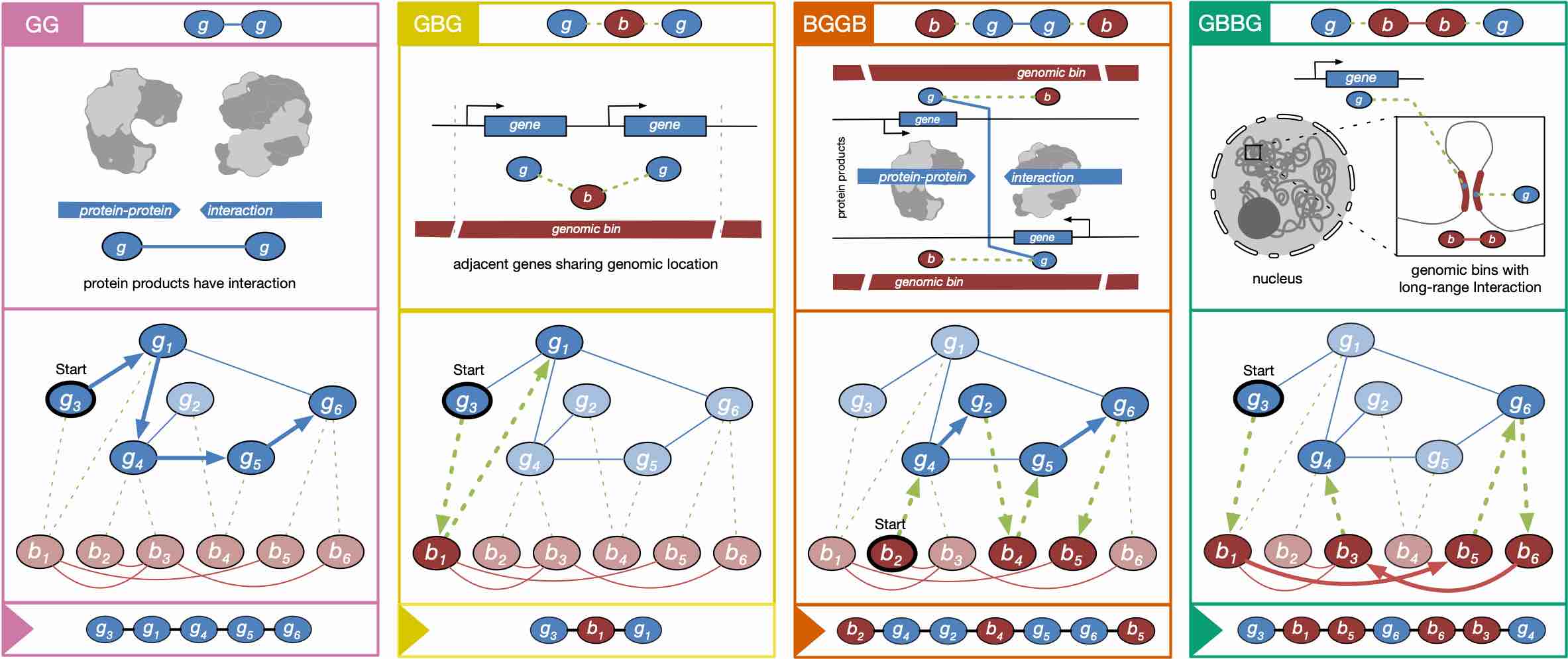
We have pre-defined 4 metapaths to reflect potential relationships among those data:
- GG (GGG...): connects two genes whose protein products have direct physical interactions, assuming that these genes are more likely to be co-expressed.
- GBG (GBGBG...): connects two genes that overlap the same genomic bin, assuming that genes adjacent in the one-dimensional genome tend to have more similar expression.
- BGGB (BGGBGGB...): connects two genomic bins that overlap genes whose protein products physically interact, assuming that these genomic bins are more likely to contain related regulatory elements such as those bound by the same transcription factors. The boundary between two occurrences of “BGGB” in a sentence also forms the “GBG” pattern, and “GG” is a sub-metapath of “BGGB”, and thus this metapath simultaneously captures three types of information.
- GBBG (GBBGBBG...): connects two genes that overlap genomic bins that are proximal in the 3D genome architecture, assuming that their expression is correlated. When a sentence is formed by merging multiple occurrences of this metapath, the boundary between two occurrences of “GBBG” also forms the “BGB” pattern, which connects two genomic bins that overlap the same gene.
Refer to our manuscript for a more detailed demonstration of GEEK on studying joint effects of different types of data on gene expression.
Minimal Demonstrative Pipeline
A minimal version (containing only processed data from GM12878, chromosome 1) of GEEK demonstrative pipeline could be downloaded here.
Dependencies
Python:
numpy
pandas
sklearn
torch
xgboost
tqdm
R:
tidyverse
reshape2
stringr
Getting started
Inputs
A pre-processed input data to GEEK is provided in this minimal pipeline, including:
- Metapath sentences (metapath folder): we have already generated sentences of 4 metapath types (GBBG, BGGB, GG, GBG) for you.
- Gene expression labels (exp_label and exp_table folder): we have binarized gene expression levels.
- 5-Folds (fold folder): we have prepared a 5-fold random split for parameter selection purpose.
- DNase Hypersensitivity Level (DNase_binary): like gene expression, they are also binarized.
Network Embedding
Network embeddings will be generated in this stage based on metapath sentences, gene expression levels (semi-supervised settings) and DNase levels.
# Generate scripts for embedding learning
Rscript 01_1_nrl_genscript.R
# Preparation for learning topologies
bash ./preprocess.allchr.sh
bash ./apply.preprocess.allchr.sh
# Preparation for learning features
bash ./semi.preprocess.allchr.sh
bash ./apply.semi.preprocess.allchr.sh
# Run GEEK Embedding Learning
bash ./nrl_cmdlist_chr1_binary.rnd.balance.sh
bash ./apply_nrl_cmdlist_chr1_binary.rnd.balance.sh
XGBoost
Best lambda pairs (\(\lambda\) values in GEEK loss function controlling the rate of gene expression semi-supervision and DNase feature) will be determined based via grid search on 5-fold cross validation for the best AUROC value in XGBoost modelling.
# Prepare for XGBoost modelling of gene expression
Rscript 02_1_xgb_assemble_csv.R
Rscript 02_2_xgb_genscript.R
Rscript 02_3_apply_xgb_assemble_csv.R
Rscript 02_4_apply_xgb_genscript.R
# Run XGBoost 5-fold parameter selection
bash ./xgb_binary_cmdlist_chr1.sh
# Collect best parameters
Rscript 03_xgb_collect.R
# Run XGBoost final modelling
bash xgb_apply_binary_cmdlist_chr1.sh
# Collect final prediction AUROC
Rscript 04_xgb_collect.R
View results
We have also prepared an R Markdown file "5_view_results.Rmd" for you to quickly see the performance. You could simply knit the file in Rstudio to see the generated "GM12878.csv" for some stats.
Whole-genome Embeddings
We have generated a set of embeddings using binarized gene expression labels of all genes, binarized DNase hypersenstivity labels, along with whole-genome PPIs and Hi-C contacts as input to GEEK, for 5 cell lines:
We hope those embeddings, as a resource, could provide some useful and interesting biological insights.
GEEK Input Data
We have prepared preprocessed data for you to further test GEEK and reproduce our results. Please refer to this page.